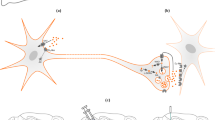
Abstract
The complex connectivity of the mammalian brain underlies its function, but understanding how interconnected brain regions interact in neural processing remains a formidable challenge. Here we address this problem by introducing a genetic probe that permits selective functional imaging of distributed neural populations defined by viral labeling techniques. The probe is an engineered enzyme that transduces cytosolic calcium dynamics of probe-expressing cells into localized hemodynamic responses that can be specifically visualized by functional magnetic resonance imaging. Using a viral vector that undergoes retrograde transport, we apply the probe to characterize a brain-wide network of presynaptic inputs to the striatum activated in a deep brain stimulation paradigm in rats. The results reveal engagement of surprisingly diverse projection sources and inform an integrated model of striatal function relevant to reward behavior and therapeutic neurostimulation approaches. Our work thus establishes a strategy for mechanistic analysis of multiregional neural systems in the mammalian brain.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Source data for Figs. 1–5 and the associated Extended Data figures are provided with this paper.
Code availability
Processing scripts used in the data analysis are available from the corresponding author on request.
References
Nakai, J., Ohkura, M. & Imoto, K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 19, 137–141 (2001).
Belliveau, J. W. et al. Functional mapping of the human visual cortex by magnetic resonance imaging. Science 254, 716–719 (1991).
Ogawa, S. et al. Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. Proc. Natl. Acad. Sci. USA 89, 5951–5955 (1992).
Hu, S., Maslov, K., Tsytsarev, V. & Wang, L. V. Functional transcranial brain imaging by optical-resolution photoacoustic microscopy. J. Biomed. Opt. 14, 040503 (2009).
Mace, E. et al. Functional ultrasound imaging of the brain. Nat. Methods 8, 662–664 (2011).
Logothetis, N. K. The underpinnings of the BOLD functional magnetic resonance imaging signal. J. Neurosci. 23, 3963–3971 (2003).
Attwell, D. et al. Glial and neuronal control of brain blood flow. Nature 468, 232–243 (2010).
Krawchuk, M. B., Ruff, C. F., Yang, X., Ross, S. E. & Vazquez, A. L. Optogenetic assessment of VIP, PV, SOM and NOS inhibitory neuron activity and cerebral blood flow regulation in mouse somato-sensory cortex. J. Cereb. Blood Flow. Metab. 40, 1427–1440 (2020).
Burke, M. & Buhrle, C. BOLD response during uncoupling of neuronal activity and CBF. Neuroimage 32, 1–8 (2006).
Stefanovic, B., Schwindt, W., Hoehn, M. & Silva, A. C. Functional uncoupling of hemodynamic from neuronal response by inhibition of neuronal nitric oxide synthase. J. Cereb. Blood Flow. Metab. 27, 741–754 (2007).
Zhou, L. & Zhu, D. Y. Neuronal nitric oxide synthase: structure, subcellular localization, regulation, and clinical implications. Nitric Oxide 20, 223–230 (2009).
Lee, S. J. & Stull, J. T. Calmodulin-dependent regulation of inducible and neuronal nitric-oxide synthase. J. Biol. Chem. 273, 27430–27437 (1998).
Brenman, J. E. et al. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and ɑ1-syntrophin mediated by PDZ domains. Cell 84, 757–767 (1996).
Schmidt, K. et al. Comparison of neuronal and endothelial isoforms of nitric oxide synthase in stably transfected HEK 293 cells. Am. J. Physiol. Heart Circ. Physiol. 281, H2053–H2061 (2001).
Shi, Z. et al. High spatial correspondence at a columnar level between activation and resting state fMRI signals and local field potentials. Proc. Natl. Acad. Sci. USA 114, 5253–5258 (2017).
Oh, S. W. et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014).
Guo, Q. et al. Whole-brain mapping of inputs to projection neurons and cholinergic interneurons in the dorsal striatum. PLoS ONE 10, e0123381 (2015).
Hunnicutt, B. J. et al. A comprehensive excitatory input map of the striatum reveals novel functional organization. eLife 5, e19103 (2016).
Mandelbaum, G. et al. Distinct cortical-thalamic-striatal circuits through the parafascicular nucleus. Neuron 102, 636–652 e637 (2019).
Stuber, G. D. & Wise, R. A. Lateral hypothalamic circuits for feeding and reward. Nat. Neurosci. 19, 198–205 (2016).
Schlaepfer, T. E., Bewernick, B. H., Kayser, S., Madler, B. & Coenen, V. A. Rapid effects of deep brain stimulation for treatment-resistant major depression. Biol. Psychiatry 73, 1204–1212 (2013).
Whiting, D. M. et al. Lateral hypothalamic area deep brain stimulation for refractory obesity: a pilot study with preliminary data on safety, body weight, and energy metabolism. J. Neurosurg. 119, 56–63 (2013).
Moisan, J. & Rompre, P. P. Electrophysiological evidence that a subset of midbrain dopamine neurons integrate the reward signal induced by electrical stimulation of the posterior mesencephalon. Brain Res. 786, 143–152 (1998).
Fenno, L. E. et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods 11, 763–772 (2014).
Li, N. & Jasanoff, A. Local and global consequences of reward-evoked striatal dopamine release. Nature 580, 239–244 (2020).
Okada, S. et al. Calcium-dependent molecular fMRI using a magnetic nanosensor. Nat. Nanotechnol. 13, 473–477 (2018).
Park, Y. G. et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol. 37, 73–83 (2018).
Biswal, B., Yetkin, F. Z., Haughton, V. M. & Hyde, J. S. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 34, 537–541 (1995).
Adamantidis, A. R. et al. Optogenetic interrogation of dopaminergic modulation of the multiple phases of reward-seeking behavior. J. Neurosci. 31, 10829–10835 (2011).
Bamford, N. S., Wightman, R. M. & Sulzer, D. Dopamine’s effects on corticostriatal synapses during reward-based behaviors. Neuron 97, 494–510 (2018).
Buxton, R. B., Uludag, K., Dubowitz, D. J. & Liu, T. T. Modeling the hemodynamic response to brain activation. Neuroimage 23, S220–S233 (2004).
Picon-Pages, P., Garcia-Buendia, J. & Munoz, F. J. Functions and dysfunctions of nitric oxide in brain. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 1949–1967 (2019).
Hillman, E. M. Coupling mechanism and significance of the BOLD signal: a status report. Annu. Rev. Neurosci. 37, 161–181 (2014).
Nippert, A. R., Biesecker, K. R. & Newman, E. A. Mechanisms mediating functional hyperemia in the brain. Neuroscientist 24, 73–83 (2018).
D’Esposito, M., Deouell, L. Y. & Gazzaley, A. Alterations in the BOLD fMRI signal with ageing and disease: a challenge for neuroimaging. Nat. Rev. Neurosci. 4, 863–872 (2003).
Jakobs, M., Fomenko, A., Lozano, A. M. & Kiening, K. L. Cellular, molecular, and clinical mechanisms of action of deep brain stimulation—a systematic review on established indications and outlook on future developments. EMBO Mol. Med. 11, e9575 (2019).
Lee, J. H. et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature 465, 788–792 (2010).
Desai, M. et al. Mapping brain networks in awake mice using combined optical neural control and fMRI. J. Neurophysiol. 105, 1393–1405 (2011).
Lima, S. Q., Hromadka, T., Znamenskiy, P. & Zador, A. M. PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS ONE 4, e6099 (2009).
Pautler, R. G. & Koretsky, A. P. Tracing odor-induced activation in the olfactory bulbs of mice using manganese-enhanced magnetic resonance imaging. Neuroimage 16, 441–448 (2002).
Yoshimura, T. et al. In vivo EPR detection and imaging of endogenous nitric oxide in lipopolysaccharide-treated mice. Nat. Biotechnol. 14, 992–994 (1996).
Reinhardt, C. J., Zhou, E. Y., Jorgensen, M. D., Partipilo, G. & Chan, J. A ratiometric acoustogenic probe for in vivo imaging of endogenous nitric oxide. J. Am. Chem. Soc. 140, 1011–1018 (2018).
Barandov, A. et al. Molecular magnetic resonance imaging of nitric oxide in biological systems. ACS Sens. 5, 1674–1682 (2020).
Wang, S., Olumolade, O. O., Sun, T., Samiotaki, G. & Konofagou, E. E. Noninvasive, neuron-specific gene therapy can be facilitated by focused ultrasound and recombinant adeno-associated virus. Gene Ther. 22, 104–110 (2015).
Deverman, B. E. et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016).
Szablowski, J. O., Lee-Gosselin, A., Lue, B., Malounda, D. & Shapiro, M. G. Acoustically targeted chemogenetics for the non-invasive control of neural circuits. Nat. Biomed. Eng. 2, 475–484 (2018).
Leithner, C. et al. Pharmacological uncoupling of activation induced increases in CBF and CMRO2. J. Cereb. Blood Flow Metab. 30, 311–322 (2010).
Qi, Y., Wang, J. K. T., McMillian, M. & Chikaraishi, D. M. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J. Neurosci. 17, 1217–1225 (1997).
Kundu, P., Inati, S. J., Evans, J. W., Luh, W. M. & Bandettini, P. A. Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. Neuroimage 60, 1759–1770 (2012).
Cox, R. W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29, 162–173 (1996).
Papp, E. A., Leergaard, T. B., Calabrese, E., Johnson, G. A. & Bjaalie, J. G. Waxholm Space atlas of the Sprague Dawley rat brain. Neuroimage 97, 374–386 (2014).
Kim, S.-Y. et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl. Acad. Sci. USA 112, E6274–E6283 (2015).
Yun, D. H. et al. Ultrafast immunostaining of organ-scale tissues for scalable proteomic phenotyping. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/660373v1 (2019).
Paxinos, G. & Watson, C. The Rat Brain in Stereotaxic Coordinates, Compact Sixth Edition (Academic Press, 2009).
Acknowledgements
This research was funded by NIH grant numbers R01 DA038642, R24 MH109081, UF1 NS107712 and U01 NS103470 and a grant from the MIT Simons Center for the Social Brain to A.J. S.G. was supported by an HHMI International Student Research Fellowship and Sheldon Razin Fellowship from the McGovern Institute for Brain Research. N.L. was supported by a Stanley Fahn Research Fellowship from the Parkinson’s Disease Foundation. M.S. was the recipient of a Marie Curie Individual Fellowship from the European Commission. T.X. was a visiting student from the Beijing University of Chinese Medicine, funded by a scholarship from the China Scholarship Council. J.I.D. was supported by the Johnson & Johnson UROP Scholars Program at MIT. We are grateful to S. Lall and B. Sabatini for comments on the manuscript, and to A. Devor, I. Wickersham and H. Sullivan for conversations. We also thank L. McLain for providing CAD cells and T. Poulos of the University of California, Irvine and P. Ortiz de Montellano at the University of California, San Francisco for providing NOS constructs. R. Neve of the Massachusetts General Hospital is acknowledged for production of HSV vectors.
Author information
Authors and Affiliations
Contributions
S.G., N.L., M.S., B.B.B. and A.J. designed the research. S.G., N.L., M.S., B.B.B., T.X., J.I.D. and U.D.S. performed the in vitro and in vivo experiments. K.X., N.D. and N.B.E. implemented the brain-clearing histology procedures under the direction of K.C. S.G., N.L., M.S. and A.J. analyzed the results. S.G., N.L. and A.J. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Alan Koretsky, Jin Hyung Lee, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 NOSTIC functionality in a neuronal cell line.
a, NOSTIC-1 fused to mCherry (cyan), as well as mCherry alone (gray), were expressed in CAD cells and stimulated with A23187 in the absence (–) or presence (+) of 100 µM 1400W; nitrite formation following NO release from the stimulated cells was measured for each condition. The effect of 1400W inhibition is significant for NOSTIC-expressing cells (paired t-test p = 0.013, n = 4) but not for control cells (p = 0.59, n = 4). b, NO production by A23187-stimulated CAD cells was visualized by staining with the fluorescent NO probe diaminofluorescein (DAF). NOSTIC-expressing cells indicated by red mCherry staining show higher levels of green DAF staining in the absence (top) than in the presence (bottom) of NOSTIC inhibition with 1400W. Scale bar = 200 µm. This experiment was performed twice.
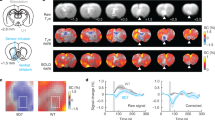
Extended Data Fig. 2 Time courses of fMRI responses to forepaw stimulation.
Mean fMRI signal time courses (% signal change) observed in the S1 forelimb region in response to stimulation of the contralateral paw (pink shaded region) in the presence vs. absence of L-NAME (left) or 1400W (right), corresponding to maps and mean response amplitudes reported in Fig. 1e. For arbitrary reasons, experiments testing effects of L-NAME and 1400W were performed with stimulation blocks of 20 s and 10 s, respectively, and differences in the response durations observed left vs. right arise from this fact.
Extended Data Fig. 3 Spatial extent of xenograft-induced hemogenetic responses.
a, Left: Anatomical MRI showing T2-weighted (T2w) contrast in the neighborhood of cell implantation for a represenative animal from the experiments of main text Fig. 2. Right: Average BOLD contrast map (color) induced by A23187 stimulation in units of percent signal change (%SC), overlayed on a T2-weighted scan. b, Cross sections of the contrast patterns through the dotted line in panel a, with solid lines denoting means and shading denoting SEM across six animals’ BOLD (red) and T2w (black) signal changes. Full width at half maximum is 1.4 ± 0.1 mm for the hemogenetic BOLD responses, indicating point-spread function comparable to or less than conventional hemodynamic fMRI.
Extended Data Fig. 4 Evaluation of HSV vectors in cell culture.
a, Western blotting was used to analyze cells transfected (Transfec.) with GFP or NOSTIC-1 (N-1), along with cells infected with HSV encoding mCherry (mC) or NOSTIC-1-IRES-mCherry (N-1/mC). Expression and size of NOSTIC-1 was confirmed using blotting with an anti-FLAG tag (α-FLAG) antibody (top). Expression and size of mCherry in the HSV infected cells was assessed using an anti-mCherry antibody (α-mCherry, middle), and anti-glyceraldehyde 3-phosphate dehydrogenase (α-GAPDH) was used as a loading control (bottom). Relevant molecular weight (MW) markers labeled at left. Expected sizes: NOSTIC-1 140 kD, mCherry 29 kD, GAPDH 37 kD. b, Griess test measurement of nitrite production in the presence of cells transfected with GFP (gray) or NOSTIC-1 (cyan) or virally transduced with mCherry or NOSTIC-1, in the presence or absence of A23187 stimulation, as indicated. Only stimulated cells expressing NOSTIC-1 show strong evidence of stimulus-dependent NO production, as indicated by the Griess test results.
Extended Data Fig. 5 LH stimulus responses in NOSTIC-expressing rats after 1400W treatment.
Average fMRI responses to LH stimulation in the +1400W condition, among 6 animals infected with NOSTIC-encoding HSV, analogous to –1400W condition data in main text Fig. 3b. Significant responses with F-test P ≤ 0.01 shown.
Extended Data Fig. 6 Time courses of responses to LH stimulation in multiple ROIs.
Average time courses of fMRI signals observed in eight ROIs in the presence and absence of 1400W, analogous to data of Fig. 3d. Pink boxes denote stimulation period. Shading denotes SEM over six animals each.
Extended Data Fig. 7 LH stimulus responses in rats treated with control HSV vectors.
Average fMRI responses to LH stimulation in the –1400W (left) and +1400W (right) conditions, among 5 animals infected with mCherry-encoding HSV, corresponding to difference maps shown in main text Fig. 3c. Significant responses with F-test P ≤ 0.01 shown.
Extended Data Fig. 8 1400W-dependent fMRI amplitudes observed in multiple ROIs.
Absolute fMRI response amplitudes observed in each of eight ROIs examined in NOSTIC and control mCherry HSV-treated animals, analogous to data presented in Fig. 3f. 1400W-dependent effects in CPu, ECx, MCx, and SN are all significant with paired t-test *P ≤ 0.05 (n = 6).
Extended Data Fig. 9 Relative changes in fMRI amplitudes upon 1400W treatment.
ROI-averaged fMRI amplitudes from the experiments of Fig. 3 were used to compute relative signal differences as 100 x (pre – post)/pre, where pre and post are the fMRI amplitudes observed before and after 1400W application in each rat. Results are shown as box plots for NOSTIC-1-treated (left, n = 6) and control mCherry-treated (right, n = 5) animals. Box plots denote median (center line), first quartiles (boxes), and full range (whiskers) over animals. NOSTIC results are significant in CPu, ECx, MCx, and SN (t-test P ≤ 0.008); control results are significant in CCx (t-test P = 0.037).
Extended Data Fig. 10 Resting-state functional connectivity to CPu.
A map of Z-transformed correlation coefficients relating resting state fMRI signal in a striatal seed region to voxels across the rest of the brain.
Supplementary information
Supplementary Information
Supplementary Table 1 and Figs. 1–6.
Source data
Source Data Fig. 1
Source data for graphs.
Source Data Fig. 2
Source data for graphs.
Source Data Fig. 3
Signal change amplitudes.
Source Data Fig. 4
Cell counting data.
Source Data Fig. 5
Source data for graphs.
Source Data Extended Data Fig. 2
Source data for time courses.
Source Data Extended Data Fig. 6
Source data for time courses.
Source Data Extended Data Fig. 8
Signal change amplitudes.
Rights and permissions
About this article
Cite this article
Ghosh, S., Li, N., Schwalm, M. et al. Functional dissection of neural circuitry using a genetic reporter for fMRI. Nat Neurosci 25, 390–398 (2022). https://doi.org/10.1038/s41593-022-01014-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01014-8
This article is cited by
-
Engineering water exchange is a safe and effective method for magnetic resonance imaging in diverse cell types
Journal of Biological Engineering (2024)
-
Structural and functional imaging of brains
Science China Chemistry (2023)